Workflow
JalilRT
2022-09-08
Last updated: 2022-09-29
Checks: 6 1
Knit directory:
NODDI-changes-by_rTMS-in_CUD/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220908) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ICVF_final_Aim1B.R | code/analysis_ICVF_final_Aim1B.R |
| ~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_OD_final_Aim1B.R | code/analysis_OD_final_Aim1B.R |
| ~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ICVF_final.R | code/analysis_ICVF_final.R |
| ~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ISOVF_final.R | code/analysis_ISOVF_final.R |
| ~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_OD_final.R | code/analysis_OD_final.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 45dcb63. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Untracked files:
Untracked: code/analysis_ICVF_final.R
Untracked: code/analysis_ICVF_final_Aim1B.R
Untracked: code/analysis_ISOVF_final.R
Untracked: code/analysis_ISOVF_final_Aim1B.R
Untracked: code/analysis_OD_final.R
Untracked: code/analysis_OD_final_Aim1B.R
Untracked: code/analysis_plots_lmm.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Workflow.Rmd) and HTML
(docs/Workflow.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 45dcb63 | JalilRT | 2022-09-29 | Updating |
| html | 31d3c57 | JalilRT | 2022-09-08 | Build site. |
| Rmd | 7542596 | JalilRT | 2022-09-08 | Updating |
Running changes in NODDI after 2 weeks rTMS
Lmer analysis for ICVF
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df_t0 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/ICVF/meanICVFinROIs_new.RDS")
subs_df_t0 <- mutate(subs_df_t0, stage = "T0")
subs_df_t1 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new_t1/ICVF_t1/meanICVFinROIs_new_t1.RDS")
subs_df_t1 <- mutate(subs_df_t1, stage = "T1")
subs_df <- rbind(subs_df_t0,subs_df_t1)
colnames(subs_df) <- c("val","rid","atlas","stage")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
subs_df <- merge(subs_df,demographics,by = "rid")
subs_df_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df$stage.x),'T',fixed=TRUE)))
subs_df_weeks <- mutate(subs_df_weeks, X3=as.numeric(X2)*2)
subs_df$stage.x <- as.numeric(subs_df_weeks$X3)
subs_df$group[subs_df$group==1] <- "sham"
subs_df$group[subs_df$group==2] <- "active"
subs_df <- mutate(subs_df,rTMS=factor(group,levels=c("sham","active")))
subs_df <- mutate(subs_df,ICVF=val)
NODDI_demo_scale <- subs_df
scale_ylab <- "ICVF"
filename_suffix <- "ICVF_lmerTest"
ylim_inf <- 0.3
ylim_sup <- 1
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
plot_lmer_probe <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
i=1
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(ICVF~stage.x*rTMS+q1_age+q2_edyears+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
plot_lmer_probe[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage.x, modx=rTMS,interval=TRUE,
x.label="weeks", y.label=scale_ylab,
main.title=k)
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/ICVF/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage.x:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL))))#,lim=c(ylim_inf,ylim_sup),
#ticks=list(at=c(ylim_sup-3*(ylim_sup-ylim_inf)/4,
# ylim_sup-2*(ylim_sup-ylim_inf)/4,
# ylim_sup-(ylim_sup-ylim_inf)/4,
# ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],3),
`Std. error`=round(coeffs[[i]][,"Std. Error"],3),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
i=i+1
}
fit_mixed_tmp <- fit_mixed_tmp %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_effects <- names(fit_mixed_tmp) %>% map(~ emmeans::emmeans(fit_mixed_tmp[[.x]],
c("stage.x","rTMS"), infer = c(T, T)) %>%
emmeans::eff_size(sigma = sigma(fit_mixed_tmp[[.x]]),
edf = df.residual(fit_mixed_tmp[[.x]]))) %>%
set_names(names(fit_mixed_tmp))
fit_effects$vmPFC2DLPFC contrast effect.size SE df lower.CL upper.CL
0 sham - 2 sham 0.458 0.316 58.3 -0.174 1.0903
0 sham - 0 active 0.831 0.550 58.3 -0.269 1.9308
0 sham - 2 active 0.180 0.555 58.3 -0.930 1.2913
2 sham - 0 active 0.373 0.556 58.3 -0.740 1.4851
2 sham - 2 active -0.278 0.564 63.4 -1.405 0.8501
0 active - 2 active -0.650 0.297 58.3 -1.244 -0.0567
sigma used for effect sizes: 0.06902
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$DLPFC2rvmPFC contrast effect.size SE df lower.CL upper.CL
0 sham - 2 sham 0.541 0.317 55.6 -0.0944 1.1764
0 sham - 0 active 1.069 0.617 55.6 -0.1678 2.3053
0 sham - 2 active 0.429 0.621 55.6 -0.8149 1.6726
2 sham - 0 active 0.528 0.621 55.6 -0.7173 1.7727
2 sham - 2 active -0.112 0.628 60.1 -1.3690 1.1447
0 active - 2 active -0.640 0.297 55.6 -1.2349 -0.0448
sigma used for effect sizes: 0.05681
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 plot_lmer_probeICV <- plot_lmer_probe %>% set_names(names(fit_mixed_tmp))Lmer analysis for OD
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df_t0 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/OD/meanODinROIs_new.RDS")
subs_df_t0 <- mutate(subs_df_t0, stage = "T0")
subs_df_t1 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new_t1/OD_t1/meanODinROIs_new_t1.RDS")
subs_df_t1 <- mutate(subs_df_t1, stage = "T1")
subs_df <- rbind(subs_df_t0,subs_df_t1)
colnames(subs_df) <- c("val","rid","atlas","stage")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
subs_df <- merge(subs_df,demographics,by = "rid")
subs_df_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df$stage.x),'T',fixed=TRUE)))
subs_df_weeks <- mutate(subs_df_weeks, X3=as.numeric(X2)*2)
subs_df$stage.x <- as.numeric(subs_df_weeks$X3)
subs_df$group[subs_df$group==1] <- "sham"
subs_df$group[subs_df$group==2] <- "active"
subs_df <- mutate(subs_df,rTMS=factor(group,levels=c("sham","active")))
subs_df <- mutate(subs_df,OD=val)
NODDI_demo_scale <- subs_df
scale_ylab <- "OD"
filename_suffix <- "OD_lmerTest"
ylim_inf <- 0
ylim_sup <- 1
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
plot_lmer_probe <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
i=1
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(OD~stage.x*rTMS+q1_age+q2_edyears+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
plot_lmer_probe[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage.x, modx=rTMS,interval=TRUE,
x.label="weeks", y.label=scale_ylab,
main.title=k)
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/OD/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage.x:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL))))#,lim=c(ylim_inf,ylim_sup),
#ticks=list(at=c(ylim_sup-3*(ylim_sup-ylim_inf)/4,
# ylim_sup-2*(ylim_sup-ylim_inf)/4,
# ylim_sup-(ylim_sup-ylim_inf)/4,
# ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],3),
`Std. error`=round(coeffs[[i]][,"Std. Error"],3),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
i=i+1
}
fit_mixed_tmp <- fit_mixed_tmp %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_effects <- names(fit_mixed_tmp) %>% map(~ emmeans::emmeans(fit_mixed_tmp[[.x]],
c("stage.x","rTMS"), infer = c(T, T)) %>%
emmeans::eff_size(sigma = sigma(fit_mixed_tmp[[.x]]),
edf = df.residual(fit_mixed_tmp[[.x]]))) %>%
set_names(names(fit_mixed_tmp))
fit_effects$DLPFC2Caud contrast effect.size SE df lower.CL upper.CL
0 sham - 2 sham 0.5642 0.317 55.1 -0.072 1.200
0 sham - 0 active 0.9443 0.630 55.1 -0.319 2.208
0 sham - 2 active 0.4767 0.635 55.1 -0.797 1.750
2 sham - 0 active 0.3800 0.635 55.1 -0.893 1.653
2 sham - 2 active -0.0876 0.643 59.4 -1.373 1.198
0 active - 2 active -0.4676 0.295 55.1 -1.059 0.124
sigma used for effect sizes: 0.01695
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$DLPFC2Thal contrast effect.size SE df lower.CL upper.CL
0 sham - 2 sham 0.5368 0.318 52.9 -0.100 1.174
0 sham - 0 active 0.4013 0.714 52.9 -1.031 1.834
0 sham - 2 active 0.0475 0.721 52.9 -1.398 1.493
2 sham - 0 active -0.1355 0.721 52.9 -1.582 1.311
2 sham - 2 active -0.4893 0.729 56.4 -1.950 0.971
0 active - 2 active -0.3538 0.295 52.9 -0.945 0.237
sigma used for effect sizes: 0.01316
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$vmPFC2DLPFC contrast effect.size SE df lower.CL upper.CL
0 sham - 2 sham 0.3512 0.315 57.1 -0.280181 0.983
0 sham - 0 active 1.1580 0.578 57.1 -0.000385 2.316
0 sham - 2 active 0.3758 0.581 57.1 -0.787524 1.539
2 sham - 0 active 0.8068 0.584 57.1 -0.362073 1.976
2 sham - 2 active 0.0246 0.589 62.0 -1.152925 1.202
0 active - 2 active -0.7821 0.299 57.1 -1.380221 -0.184
sigma used for effect sizes: 0.04831
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 plot_lmer_probeOD <- plot_lmer_probe %>% set_names(names(fit_mixed_tmp))Lmer analysis for ISOVF
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df_t0 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/ISOVF/meanISOVFinROIs_new.RDS")
subs_df_t0 <- mutate(subs_df_t0, stage = "T0")
subs_df_t1 <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new_t1/ISOVF_t1/meanISOVFinROIs_new_t1.RDS")
subs_df_t1 <- mutate(subs_df_t1, stage = "T1")
subs_df <- rbind(subs_df_t0,subs_df_t1)
colnames(subs_df) <- c("val","rid","atlas","stage")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
subs_df <- merge(subs_df,demographics,by = "rid")
subs_df_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df$stage.x),'T',fixed=TRUE)))
subs_df_weeks <- mutate(subs_df_weeks, X3=as.numeric(X2)*2)
subs_df$stage.x <- as.numeric(subs_df_weeks$X3)
subs_df$group[subs_df$group==1] <- "sham"
subs_df$group[subs_df$group==2] <- "active"
subs_df <- mutate(subs_df,rTMS=factor(group,levels=c("sham","active")))
subs_df <- mutate(subs_df,ISOVF=val)
NODDI_demo_scale <- subs_df
scale_ylab <- "CSF volume fraction (ISOVF)"
filename_suffix <- "ISOVF_lmerTest"
ylim_inf <- 0
ylim_sup <- 1
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
plot_lmer_probe <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
i=1
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(ISOVF~stage.x*rTMS+q1_age+q2_edyears+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
plot_lmer_probe[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage.x, modx=rTMS,interval=TRUE,
x.label="weeks", y.label=scale_ylab,
main.title=k)
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("C:/Users/vissa/Documents/Universitaeten/TEC/Cl?nicas/2021/UNAM/TMS/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/ISOVF/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
print(e)
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage.x:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL))))#,lim=c(ylim_inf,ylim_sup),
#ticks=list(at=c(ylim_sup-3*(ylim_sup-ylim_inf)/4,
# ylim_sup-2*(ylim_sup-ylim_inf)/4,
# ylim_sup-(ylim_sup-ylim_inf)/4,
# ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],3),
`Std. error`=round(coeffs[[i]][,"Std. Error"],3),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
i=i+1
} model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.3007132 0.3096426 0.3174559 0.3252692 0.3341986
q2_edyears effect
q2_edyears
8 11 14 16 19
0.3332848 0.3249795 0.3166743 0.3111374 0.3028322
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.2998029 0.3342144
0.5 0.2999014 0.3349548
1 0.3000000 0.3356952
1.5 0.3000985 0.3364356
2 0.3001970 0.3371760
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.1979832 0.2146103 0.2291590 0.2437077 0.2603348
q2_edyears effect
q2_edyears
8 11 14 16 19
0.2502507 0.2397022 0.2291538 0.2221214 0.2115730
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.2157556 0.2436727
0.5 0.2159642 0.2450025
1 0.2161727 0.2463322
1.5 0.2163813 0.2476620
2 0.2165899 0.2489918
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.1442666 0.1645274 0.1822556 0.1999838 0.2202447
q2_edyears effect
q2_edyears
8 11 14 16 19
0.1865977 0.1862711 0.1859446 0.1857269 0.1854003
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.1809495 0.1893512
0.5 0.1807285 0.1901189
1 0.1805075 0.1908866
1.5 0.1802865 0.1916543
2 0.1800655 0.1924221
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.2266485 0.2555826 0.2809000 0.3062173 0.3351514
q2_edyears effect
q2_edyears
8 11 14 16 19
0.2738383 0.2811505 0.2884626 0.2933374 0.3006495
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.2919627 0.2776425
0.5 0.2920182 0.2796484
1 0.2920737 0.2816543
1.5 0.2921293 0.2836602
2 0.2921848 0.2856661
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.1947669 0.2128159 0.2286088 0.2444016 0.2624506
q2_edyears effect
q2_edyears
8 11 14 16 19
0.2433918 0.2366992 0.2300066 0.2255449 0.2188523
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.2226286 0.2387205
0.5 0.2222049 0.2397896
1 0.2217811 0.2408587
1.5 0.2213573 0.2419277
2 0.2209336 0.2429968
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.1305534 0.1424569 0.1528724 0.1632880 0.1751914
q2_edyears effect
q2_edyears
8 11 14 16 19
0.1575058 0.1560926 0.1546795 0.1537374 0.1523242
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.1449543 0.1623531
0.5 0.1463984 0.1619035
1 0.1478424 0.1614540
1.5 0.1492865 0.1610044
2 0.1507306 0.1605548
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.3983583 0.4068808 0.4143379 0.4217951 0.4303176
q2_edyears effect
q2_edyears
8 11 14 16 19
0.4383461 0.4251991 0.4120520 0.4032873 0.3901402
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.3958560 0.4317568
0.5 0.3954388 0.4328926
1 0.3950216 0.4340285
1.5 0.3946044 0.4351643
2 0.3941873 0.4363001
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.2983354 0.3155563 0.3306246 0.3456928 0.3629137
q2_edyears effect
q2_edyears
8 11 14 16 19
0.3669182 0.3475188 0.3281194 0.3151865 0.2957872
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.3177543 0.3445106
0.5 0.3174834 0.3464558
1 0.3172124 0.3484010
1.5 0.3169414 0.3503462
2 0.3166705 0.3522914
model: ISOVF ~ stage.x * rTMS + q1_age + q2_edyears
q1_age effect
q1_age
18 26 33 40 48
0.1160231 0.1290268 0.1404050 0.1517833 0.1647869
q2_edyears effect
q2_edyears
8 11 14 16 19
0.1371555 0.1404863 0.1438170 0.1460375 0.1493682
stage.x*rTMS effect
rTMS
stage.x sham active
0 0.1454649 0.1358732
0.5 0.1447790 0.1390039
1 0.1440930 0.1421346
1.5 0.1434070 0.1452654
2 0.1427211 0.1483961Running baseline NODDI as a predictor of clinical outcome after rTMS
Lmer analysis for ICVF
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
#library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/ICVF/meanICVFinROIs_new.RDS")
VAS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/VAS.csv")
CCQ_N <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/CCQ-N.csv")
Inventory <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/Inventory.csv")
BIS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/BIS-11.csv")
colnames(subs_df) <- c("val","rid","atlas")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
VAS_t0t1 <- filter(VAS,(stage=='T0'|stage=='T1'))#&group==2)
CCQ_N_t0t1 <- filter(CCQ_N,(stage=='T0'|stage=='T1'))#&group==2)
Inventory_t0t1 <- filter(Inventory,(stage=='T0'|stage=='T1'))#&group==2)
BIS_t0t1 <- filter(BIS,(stage=='T0'|stage=='T1'))#&group==2)
create_pivot_table <- function(table_name,score_name){
pt <- PivotTable$new()
pt$addData(table_name)
pt$addColumnDataGroups("stage")
pt$addRowDataGroups("rid")
pt$defineCalculation(calculationName=score_name,summariseExpression=paste("sum(",score_name,")",sep = "") )
pt$renderPivot()
table_VAS_t0t1 <- pt$asDataFrame()
table_VAS_t0t1 <- mutate(table_VAS_t0t1, rid = rownames(table_VAS_t0t1))
table_VAS_t0t1 <- subset(table_VAS_t0t1,select = -c(Total))
table_VAS_t0t1 <- table_VAS_t0t1[!(row.names(table_VAS_t0t1) %in% "Total"), ]
return(table_VAS_t0t1)
}
#save.image(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
#load(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
#subs_df <- merge(subs_df, table_cocmet_COI_EXC_VAS_CCQ)
#subs_df <- mutate(subs_df,responder_final=factor(Responder,levels = c(FALSE,TRUE)))
subs_df <- merge(subs_df,demographics,by = "rid")
scale_prep <- function(NODDImetrics_AND_demo,scale_to_merge,varname){
subs_df_t1 <- NODDImetrics_AND_demo
subs_df_t1$stage <- "T1"
#subs_df_t1$val <- NA
subs_df_t0t1 <- rbind(NODDImetrics_AND_demo,subs_df_t1)
subs_df_CCQ_N <- merge(subs_df_t0t1,scale_to_merge, by =c("rid","stage"),all.x=TRUE)
subs_df_CCQ_N_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df_CCQ_N$stage),'T',fixed=TRUE)))
subs_df_CCQ_N_weeks <- mutate(subs_df_CCQ_N_weeks, X3=as.numeric(X2)*2)
subs_df_CCQ_N$stage <- as.numeric(subs_df_CCQ_N_weeks$X3)
#subs_df_VAS2 <- mutate(subs_df_VAS2,stage=factor(stage,levels = c(0,2)))
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==1] <- "sham"
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==2] <- "active"
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,rTMS=factor(group.x,levels=c("sham","active")))
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,ICVF=val)
#subs_df_CCQ_N <- mutate(subs_df_VAS2,`ICVF-Z-score`=scale(val))
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=group.x-1)
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=factor(rTMS,levels = c(0,1)))
#subs_df_VAS2 <- mutate(subs_df_VAS2,val=val-min(val))
#subs_df_CCQ_N <- mutate(subs_df_CCQ_N,scale=eval(varname))
names(subs_df_CCQ_N)[names(subs_df_CCQ_N) == varname] <- 'scale'
return(subs_df_CCQ_N)
}
subs_df_VAS <- scale_prep(subs_df,VAS_t0t1,"vas")
#plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
NODDI_demo_scale<-select(subs_df_VAS, rid,atlas,rTMS,stage,q1_age,q2_edyears,ICVF,scale)
NODDI_demo_scale <- arrange(NODDI_demo_scale,atlas,rid,stage, rTMS)
scale_ylab <- "Craving VAS score"
filename_suffix <- "VAS_lmerTest"
ylim_inf <- 0
ylim_sup <- 10
# subs_df_CCQ_N <- scale_prep(subs_df,CCQ_N_t0t1,"ccq_n")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-subs_df_CCQ_N
# scale_ylab <- "craving CCQnow"
# filename_suffix <- "CCQnow_lmerTest"
# ylim_inf <- 0
# ylim_sup <- 250
#
# subs_df_BIStot <- scale_prep(subs_df,BIS_t0t1,"tot_score")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-subs_df_BIStot
# scale_ylab <- "total BIS11"
# filename_suffix <- "BIStot_lmerTest"
# ylim_inf <- 0#min(NODDI_demo_scale$scale)
# ylim_sup <- 100#max(NODDI_demo_scale$scale)
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
#p_values <- c()
i=1
plot_lmer_gg <- list()
plot_lmer_gg2 <- list()
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(scale~stage*ICVF*rTMS+q1_age*ICVF+q2_edyears*ICVF+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
#plot_lmer[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage, modx=rTMS,mod2=ICVF,interval=TRUE,
# mod2.values=quantile(tmp$val, c(.05, .5, .95)) ,x.label="weeks",
# y.label=scale_ylab, main.title=paste(k,"\npercentile of neurite density (ICVF)",sep=""))
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/ICVF/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
levs <- e$`stage:ICVF:rTMS` %>% as_tibble() %>% select(ICVF) %>%
c() %>% unlist()%>% as_factor() %>% levels()
plot_lmer_gg[[i]] <- e$`stage:ICVF:rTMS` %>% as_tibble() %>%
mutate(ICVF = case_when(ICVF==levs[1]~paste0("ICVF = ",levs[1]),
ICVF==levs[2]~paste0("ICVF = ",levs[2]),
ICVF==levs[3]~paste0("ICVF = ",levs[3]),
ICVF==levs[4]~paste0("ICVF = ",levs[4]),
ICVF==levs[5]~paste0("ICVF = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = rTMS)) +
geom_line() + facet_wrap(~ICVF)+
geom_ribbon(aes(ymin=lower, ymax=upper,fill=rTMS), colour=NA,alpha=0.1) +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
scale_color_manual(values = c("#ff7b00","#49b7fc")) +
theme(text = element_text(size = 16))
plot_lmer_gg2[[i]] <- e$`stage:ICVF:rTMS` %>% as_tibble() %>%
mutate(ICVF = case_when(ICVF==levs[1]~paste0("ICVF = ",levs[1]),
ICVF==levs[2]~paste0("ICVF = ",levs[2]),
ICVF==levs[3]~paste0("ICVF = ",levs[3]),
ICVF==levs[4]~paste0("ICVF = ",levs[4]),
ICVF==levs[5]~paste0("ICVF = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = ICVF)) +
geom_ribbon(aes(ymin=lower, ymax=upper,fill=ICVF), colour=NA,alpha=0.08) +
geom_line() + facet_wrap(~rTMS) +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
scale_color_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
scale_fill_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
theme(text = element_text(size = 16),legend.title = element_blank())
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage:ICVF:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL),lim=c(ylim_inf,ylim_sup),
ticks=list(at=c(ylim_sup-(ylim_sup-ylim_inf)/2,
ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],2),
`Std. error`=round(coeffs[[i]][,"Std. Error"],2),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
#p_values <- c(p_values,coeffs[[i]][,"Pr(>|t|)"])
i=i+1
}
plot_lmer_gg <- plot_lmer_gg %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
plot_lmer_gg2 <- plot_lmer_gg2 %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_mixed_tmp <- fit_mixed_tmp %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_effects <- names(fit_mixed_tmp) %>% map(~ emmeans::emmeans(fit_mixed_tmp[[.x]],
c("stage","ICVF","rTMS"), infer = c(T, T)) %>%
emmeans::eff_size(sigma = sigma(fit_mixed_tmp[[.x]]),
edf = df.residual(fit_mixed_tmp[[.x]]))) %>%
set_names(names(fit_mixed_tmp))
fit_effects$Caud2Medulla contrast effect.size SE df
0 0.815147662338002 sham - 2 0.815147662338002 sham 0.164 0.314 59.0
0 0.815147662338002 sham - 0 0.815147662338002 active -0.445 0.464 58.7
0 0.815147662338002 sham - 2 0.815147662338002 active 0.893 0.478 59.0
2 0.815147662338002 sham - 0 0.815147662338002 active -0.609 0.479 58.7
2 0.815147662338002 sham - 2 0.815147662338002 active 0.729 0.491 65.2
0 0.815147662338002 active - 2 0.815147662338002 active 1.338 0.311 58.7
lower.CL upper.CL
-0.464 0.793
-1.373 0.483
-0.064 1.850
-1.567 0.349
-0.251 1.708
0.716 1.959
sigma used for effect sizes: 1.886
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$Caud2Palli contrast effect.size SE df
0 0.81940682680827 sham - 2 0.81940682680827 sham 0.199 0.314 58.8
0 0.81940682680827 sham - 0 0.81940682680827 active -0.426 0.463 58.6
0 0.81940682680827 sham - 2 0.81940682680827 active 0.937 0.478 58.8
2 0.81940682680827 sham - 0 0.81940682680827 active -0.625 0.477 58.6
2 0.81940682680827 sham - 2 0.81940682680827 active 0.738 0.489 64.8
0 0.81940682680827 active - 2 0.81940682680827 active 1.363 0.311 58.6
lower.CL upper.CL
-0.4289 0.826
-1.3523 0.500
-0.0201 1.894
-1.5789 0.329
-0.2379 1.714
0.7401 1.986
sigma used for effect sizes: 1.842
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$Thal2Medulla contrast effect.size SE df
0 0.76162621353232 sham - 2 0.76162621353232 sham 0.173 0.315 58.0
0 0.76162621353232 sham - 0 0.76162621353232 active -0.499 0.476 57.9
0 0.76162621353232 sham - 2 0.76162621353232 active 0.822 0.489 58.0
2 0.76162621353232 sham - 0 0.76162621353232 active -0.672 0.489 57.9
2 0.76162621353232 sham - 2 0.76162621353232 active 0.649 0.499 63.9
0 0.76162621353232 active - 2 0.76162621353232 active 1.321 0.310 57.9
lower.CL upper.CL
-0.457 0.803
-1.453 0.454
-0.156 1.800
-1.651 0.306
-0.347 1.645
0.700 1.942
sigma used for effect sizes: 1.894
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$Thal2Palli contrast effect.size SE df
0 0.776816694724299 sham - 2 0.776816694724299 sham 0.212 0.314 56.5
0 0.776816694724299 sham - 0 0.776816694724299 active -0.495 0.499 56.4
0 0.776816694724299 sham - 2 0.776816694724299 active 0.935 0.512 56.5
2 0.776816694724299 sham - 0 0.776816694724299 active -0.707 0.512 56.4
2 0.776816694724299 sham - 2 0.776816694724299 active 0.723 0.522 62.2
0 0.776816694724299 active - 2 0.776816694724299 active 1.430 0.315 56.4
lower.CL upper.CL
-0.4174 0.841
-1.4942 0.503
-0.0913 1.960
-1.7318 0.317
-0.3196 1.765
0.8000 2.060
sigma used for effect sizes: 1.809
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 Lmer analysis for OD
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
#library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/OD/meanODinROIs_new.RDS")
VAS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/VAS.csv")
CCQ_N <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/CCQ-N.csv")
Inventory <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/Inventory.csv")
BIS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/BIS-11.csv")
colnames(subs_df) <- c("val","rid","atlas")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
VAS_t0t1 <- filter(VAS,(stage=='T0'|stage=='T1'))#&group==2)
CCQ_N_t0t1 <- filter(CCQ_N,(stage=='T0'|stage=='T1'))#&group==2)
Inventory_t0t1 <- filter(Inventory,(stage=='T0'|stage=='T1'))#&group==2)
BIS_t0t1 <- filter(BIS,(stage=='T0'|stage=='T1'))#&group==2)
create_pivot_table <- function(table_name,score_name){
pt <- PivotTable$new()
pt$addData(table_name)
pt$addColumnDataGroups("stage")
pt$addRowDataGroups("rid")
pt$defineCalculation(calculationName=score_name,summariseExpression=paste("sum(",score_name,")",sep = "") )
pt$renderPivot()
table_VAS_t0t1 <- pt$asDataFrame()
table_VAS_t0t1 <- mutate(table_VAS_t0t1, rid = rownames(table_VAS_t0t1))
table_VAS_t0t1 <- subset(table_VAS_t0t1,select = -c(Total))
table_VAS_t0t1 <- table_VAS_t0t1[!(row.names(table_VAS_t0t1) %in% "Total"), ]
return(table_VAS_t0t1)
}
#save.image(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
#load(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
#subs_df <- merge(subs_df, table_cocmet_COI_EXC_VAS_CCQ)
#subs_df <- mutate(subs_df,responder_final=factor(Responder,levels = c(FALSE,TRUE)))
subs_df <- merge(subs_df,demographics,by = "rid")
scale_prep <- function(NODDImetrics_AND_demo,scale_to_merge,varname){
subs_df_t1 <- NODDImetrics_AND_demo
subs_df_t1$stage <- "T1"
#subs_df_t1$val <- NA
subs_df_t0t1 <- rbind(NODDImetrics_AND_demo,subs_df_t1)
subs_df_CCQ_N <- merge(subs_df_t0t1,scale_to_merge, by =c("rid","stage"),all.x=TRUE)
subs_df_CCQ_N_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df_CCQ_N$stage),'T',fixed=TRUE)))
subs_df_CCQ_N_weeks <- mutate(subs_df_CCQ_N_weeks, X3=as.numeric(X2)*2)
subs_df_CCQ_N$stage <- as.numeric(subs_df_CCQ_N_weeks$X3)
#subs_df_VAS2 <- mutate(subs_df_VAS2,stage=factor(stage,levels = c(0,2)))
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==1] <- "sham"
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==2] <- "active"
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,rTMS=factor(group.x,levels=c("sham","active")))
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,OD=val)
#subs_df_CCQ_N <- mutate(subs_df_VAS2,`OD-Z-score`=scale(val))
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=group.x-1)
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=factor(rTMS,levels = c(0,1)))
#subs_df_VAS2 <- mutate(subs_df_VAS2,val=val-min(val))
#subs_df_CCQ_N <- mutate(subs_df_CCQ_N,scale=eval(varname))
names(subs_df_CCQ_N)[names(subs_df_CCQ_N) == varname] <- 'scale'
return(subs_df_CCQ_N)
}
# subs_df_VAS <- scale_prep(subs_df,VAS_t0t1,"vas")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-select(subs_df_VAS, rid,atlas,rTMS,stage,q1_age,q2_edyears,OD,scale)
# NODDI_demo_scale <- arrange(NODDI_demo_scale,atlas,rid,stage, rTMS)
# scale_ylab <- "craving VAS"
# filename_suffix <- "VAS_lmerTest"
# ylim_inf <- 0
# ylim_sup <- 10
#
# subs_df_CCQ_N <- scale_prep(subs_df,CCQ_N_t0t1,"ccq_n")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-subs_df_CCQ_N
# scale_ylab <- "craving CCQnow"
# filename_suffix <- "CCQnow_lmerTest"
# ylim_inf <- 0
# ylim_sup <- 250
subs_df_BIStot <- scale_prep(subs_df,BIS_t0t1,"tot_score")
#plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
NODDI_demo_scale<-subs_df_BIStot
scale_ylab <- "Total BIS-11 score"
filename_suffix <- "BIStot_lmerTest"
ylim_inf <- 0#min(NODDI_demo_scale$scale)
ylim_sup <- 100#max(NODDI_demo_scale$scale)
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
i=1
plot_lmer_gg <- list()
plot_lmer_gg2 <- list()
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(scale~stage*OD*rTMS+q1_age*OD+q2_edyears*OD+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
#plot_lmer[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage, modx=rTMS,mod2=OD,interval=TRUE,
# mod2.values=quantile(tmp$val, c(.05, .5, .95)) ,x.label="weeks",
# y.label=scale_ylab, main.title=paste(k,"\npercentile of neurite density (OD)",sep=""))
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/OD/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
levs <- e$`stage:OD:rTMS` %>% as_tibble() %>% select(OD) %>%
c() %>% unlist()%>% as_factor() %>% levels()
plot_lmer_gg[[i]] <- e$`stage:OD:rTMS` %>% as_tibble() %>%
mutate(OD = case_when(OD==levs[1]~paste0("OD = ",levs[1]),
OD==levs[2]~paste0("OD = ",levs[2]),
OD==levs[3]~paste0("OD = ",levs[3]),
OD==levs[4]~paste0("OD = ",levs[4]),
OD==levs[5]~paste0("OD = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = rTMS)) +
geom_line() + facet_wrap(~OD)+
geom_ribbon(aes(ymin=lower, ymax=upper,fill=rTMS), colour=NA,alpha=0.08) +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
scale_color_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
scale_fill_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
theme(text = element_text(size = 16))
plot_lmer_gg2[[i]] <- e$`stage:OD:rTMS` %>% as_tibble() %>%
mutate(OD = case_when(OD==levs[1]~paste0("OD = ",levs[1]),
OD==levs[2]~paste0("OD = ",levs[2]),
OD==levs[3]~paste0("OD = ",levs[3]),
OD==levs[4]~paste0("OD = ",levs[4]),
OD==levs[5]~paste0("OD = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = OD)) +
geom_ribbon(aes(ymin=lower, ymax=upper,fill=OD), colour=NA,alpha=0.1) +
geom_line() + facet_wrap(~rTMS)+
ggthemes::scale_color_wsj() +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
theme(text = element_text(size = 16),legend.title = element_blank())
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage:OD:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL),lim=c(ylim_inf,ylim_sup),
ticks=list(at=c(ylim_sup-(ylim_sup-ylim_inf)/2,
ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],2),
`Std. error`=round(coeffs[[i]][,"Std. Error"],2),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
#p_values <- c(p_values,coeffs[[i]][,"Pr(>|t|)"])
i=i+1
}
plot_lmer_gg <- plot_lmer_gg %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
plot_lmer_gg2 <- plot_lmer_gg2 %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_mixed_tmp <- fit_mixed_tmp %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_effects <- names(fit_mixed_tmp) %>% map(~ emmeans::emmeans(fit_mixed_tmp[[.x]],
c("stage","OD","rTMS"), infer = c(T, T)) %>%
emmeans::eff_size(sigma = sigma(fit_mixed_tmp[[.x]]),
edf = df.residual(fit_mixed_tmp[[.x]]))) %>%
set_names(names(fit_mixed_tmp))
fit_effects$DLPFC2rvmPFC contrast effect.size SE df
0 0.37951551078487 sham - 2 0.37951551078487 sham 0.275 0.325 51.9
0 0.37951551078487 sham - 0 0.37951551078487 active -0.310 0.587 51.4
0 0.37951551078487 sham - 2 0.37951551078487 active 1.103 0.602 51.9
2 0.37951551078487 sham - 0 0.37951551078487 active -0.585 0.597 51.4
2 0.37951551078487 sham - 2 0.37951551078487 active 0.827 0.608 56.0
0 0.37951551078487 active - 2 0.37951551078487 active 1.412 0.319 51.4
lower.CL upper.CL
-0.377 0.927
-1.488 0.868
-0.106 2.312
-1.783 0.613
-0.391 2.045
0.772 2.053
sigma used for effect sizes: 8.7
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 Lmer analysis for ISOVF
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
#library(interactions)
#library(sjPlot)
library(lmerTest)
subs_df <- readRDS("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/ROIs_new/ISOVF/meanISOVFinROIs_new.RDS")
VAS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/VAS.csv")
CCQ_N <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/CCQ-N.csv")
Inventory <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/Inventory.csv")
BIS <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/BIS-11.csv")
colnames(subs_df) <- c("val","rid","atlas")
subs_ids <- data.frame(do.call('rbind', strsplit(as.character(subs_df$rid),'sub-',fixed=TRUE)))
subs_df$rid <- as.numeric(subs_ids$X2)
VAS_t0t1 <- filter(VAS,(stage=='T0'|stage=='T1'))#&group==2)
CCQ_N_t0t1 <- filter(CCQ_N,(stage=='T0'|stage=='T1'))#&group==2)
Inventory_t0t1 <- filter(Inventory,(stage=='T0'|stage=='T1'))#&group==2)
BIS_t0t1 <- filter(BIS,(stage=='T0'|stage=='T1'))#&group==2)
create_pivot_table <- function(table_name,score_name){
pt <- PivotTable$new()
pt$addData(table_name)
pt$addColumnDataGroups("stage")
pt$addRowDataGroups("rid")
pt$defineCalculation(calculationName=score_name,summariseExpression=paste("sum(",score_name,")",sep = "") )
pt$renderPivot()
table_VAS_t0t1 <- pt$asDataFrame()
table_VAS_t0t1 <- mutate(table_VAS_t0t1, rid = rownames(table_VAS_t0t1))
table_VAS_t0t1 <- subset(table_VAS_t0t1,select = -c(Total))
table_VAS_t0t1 <- table_VAS_t0t1[!(row.names(table_VAS_t0t1) %in% "Total"), ]
return(table_VAS_t0t1)
}
#save.image(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
#load(file = "~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/pivot_tables.Rdata")
demographics <- read_csv("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/Addimex_clinical_Feb20/csv/DEMOGRAPHIC.csv")
subs_df <- merge(subs_df,demographics,by = "rid")
scale_prep <- function(NODDImetrics_AND_demo,scale_to_merge,varname){
subs_df_t1 <- NODDImetrics_AND_demo
subs_df_t1$stage <- "T1"
#subs_df_t1$val <- NA
subs_df_t0t1 <- rbind(NODDImetrics_AND_demo,subs_df_t1)
subs_df_CCQ_N <- merge(subs_df_t0t1,scale_to_merge, by =c("rid","stage"),all.x=TRUE)
subs_df_CCQ_N_weeks <- data.frame(do.call('rbind', strsplit(as.character(subs_df_CCQ_N$stage),'T',fixed=TRUE)))
subs_df_CCQ_N_weeks <- mutate(subs_df_CCQ_N_weeks, X3=as.numeric(X2)*2)
subs_df_CCQ_N$stage <- as.numeric(subs_df_CCQ_N_weeks$X3)
#subs_df_VAS2 <- mutate(subs_df_VAS2,stage=factor(stage,levels = c(0,2)))
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==1] <- "sham"
subs_df_CCQ_N$group.x[subs_df_CCQ_N$group.x==2] <- "active"
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,rTMS=factor(group.x,levels=c("sham","active")))
subs_df_CCQ_N <- mutate(subs_df_CCQ_N,ISOVF=val)
#subs_df_CCQ_N <- mutate(subs_df_VAS2,`ISOVF-Z-score`=scale(val))
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=group.x-1)
#subs_df_VAS2 <- mutate(subs_df_VAS2,rTMS=factor(rTMS,levels = c(0,1)))
#subs_df_VAS2 <- mutate(subs_df_VAS2,val=val-min(val))
#subs_df_CCQ_N <- mutate(subs_df_CCQ_N,scale=eval(varname))
names(subs_df_CCQ_N)[names(subs_df_CCQ_N) == varname] <- 'scale'
return(subs_df_CCQ_N)
}
# subs_df_VAS <- scale_prep(subs_df,VAS_t0t1,"vas")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-select(subs_df_VAS, rid,atlas,rTMS,stage,q1_age,q2_edyears,ISOVF,scale)
# NODDI_demo_scale <- arrange(NODDI_demo_scale,atlas,rid,stage, rTMS)
# scale_ylab <- "craving VAS"
# filename_suffix <- "VAS_lmerTest"
# ylim_inf <- 0
# ylim_sup <- 10
#
# subs_df_CCQ_N <- scale_prep(subs_df,CCQ_N_t0t1,"ccq_n")
# #plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
# NODDI_demo_scale<-subs_df_CCQ_N
# scale_ylab <- "craving CCQnow"
# filename_suffix <- "CCQnow_lmerTest"
# ylim_inf <- 0
# ylim_sup <- 250
subs_df_BIStot <- scale_prep(subs_df,BIS_t0t1,"tot_score")
#plotANDsavetables(subs_df_CCQ_N,"craving CCQnow","CCQnow",0,200)
NODDI_demo_scale<-subs_df_BIStot
scale_ylab <- "Total BIS-11 score"
filename_suffix <- "BIStot_lmerTest"
ylim_inf <- 0#min(NODDI_demo_scale$scale)
ylim_sup <- 100#max(NODDI_demo_scale$scale)
#plotANDsavetables <- function(NODDI_demo_scale,scale_ylab,filename_suffix,ylim_inf,ylim_sup){
# library(lme4)
plot_lmer <- list()
fit_mixed_tmp <- list()
coeffs <- list()
coeffs_tib <- list()
i=1
plot_lmer_gg <- list()
plot_lmer_gg2 <- list()
for(k in sort(unique(NODDI_demo_scale$atlas))) {
tmp <- filter(NODDI_demo_scale, atlas == k)
fit_mixed_tmp[[i]] <- lmer(scale~stage*ISOVF*rTMS+q1_age*ISOVF+q2_edyears*ISOVF+(1|rid),data=tmp)
#summary(fit_mixed_tmp[[i]])
#confint(fit_mixed_tmp[i])
# ranef(fit_mixed_tmp)
# coef(fit_mixed_tmp)
# predict_with_re <- predict(fit_mixed_tmp)
#Anova(fit_mixed_tmp[[i]])
#plot_lmer[[i]] <- probe_interaction(fit_mixed_tmp[[i]],pred=stage, modx=rTMS,mod2=ISOVF,interval=TRUE,
# mod2.values=quantile(tmp$val, c(.05, .5, .95)) ,x.label="weeks",
# y.label=scale_ylab, main.title=paste(k,"\npercentile of neurite density (ISOVF)",sep=""))
#tab_model(fit_mixed_tmp[[i]],p.val = "kr", show.df = TRUE)#,
#file=paste("~/Documents/Psilantro/NODDI_tms/postprocess_HARDI/NODDI_all/VAS_CCQnow_InstantView_plots/continuous/ISOVF/",
# filename_suffix,"/", k,"_",filename_suffix,".html",sep = ""))
e <- allEffects(fit_mixed_tmp[[i]])
levs <- e$`stage:ISOVF:rTMS` %>% as_tibble() %>% select(ISOVF) %>%
c() %>% unlist()%>% as_factor() %>% levels()
plot_lmer_gg[[i]] <- e$`stage:ISOVF:rTMS` %>% as_tibble() %>%
mutate(ISOVF = case_when(ISOVF==levs[1]~paste0("ISOVF = ",levs[1]),
ISOVF==levs[2]~paste0("ISOVF = ",levs[2]),
ISOVF==levs[3]~paste0("ISOVF = ",levs[3]),
ISOVF==levs[4]~paste0("ISOVF = ",levs[4]),
ISOVF==levs[5]~paste0("ISOVF = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = rTMS)) +
geom_line() + facet_wrap(~ISOVF)+
geom_ribbon(aes(ymin=lower, ymax=upper,fill=rTMS), colour=NA,alpha=0.08) +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
scale_color_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
scale_fill_manual(values = c("#1B9E77","#D95F02","#7570B3","#E7298A","#4DBBD5FF")) +
theme(text = element_text(size = 16))
plot_lmer_gg2[[i]] <- e$`stage:ISOVF:rTMS` %>% as_tibble() %>%
mutate(ISOVF = case_when(ISOVF==levs[1]~paste0("ISOVF = ",levs[1]),
ISOVF==levs[2]~paste0("ISOVF = ",levs[2]),
ISOVF==levs[3]~paste0("ISOVF = ",levs[3]),
ISOVF==levs[4]~paste0("ISOVF = ",levs[4]),
ISOVF==levs[5]~paste0("ISOVF = ",levs[5])) ) %>%
ggplot(aes(x = stage, y = fit, colour = ISOVF)) +
geom_ribbon(aes(ymin=lower, ymax=upper,fill=ISOVF), colour=NA,alpha=0.1) +
geom_line() + facet_wrap(~rTMS)+
ggthemes::scale_color_wsj() +
theme_bw() + labs(x="Weeks", y = scale_ylab) +
theme(text = element_text(size = 16),legend.title = element_blank())
#plot(e)
plot_lmer[[i]] <-
plot(e$`stage:ISOVF:rTMS`,multiline=TRUE,confint=TRUE,ci.style="bands",main=k,
lattice = list(key.args=list(space="none")),
axes = list(x=list(stage=list(lab=list(label=NULL),lim=c(0,2),ticks=list(at=c(0,2)))),
y=list(lab=list(label=NULL),lim=c(ylim_inf,ylim_sup),
ticks=list(at=c(ylim_sup-(ylim_sup-ylim_inf)/2,
ylim_sup)))))
summa <- summary(fit_mixed_tmp[[i]])
coeffs_w_names <- summa$coefficients
coeffs[[i]] <- coeffs_w_names
row.names(coeffs[[i]]) <- NULL
coeffs_tib[[i]] <- dplyr::tibble(Estimate=round(coeffs[[i]][,"Estimate"],2),
`Std. error`=round(coeffs[[i]][,"Std. Error"],2),
df=round(coeffs[[i]][,"df"],2),
`p-value`=round(coeffs[[i]][,"Pr(>|t|)"],3))
#p_values <- c(p_values,coeffs[[i]][,"Pr(>|t|)"])
i=i+1
}
plot_lmer_gg <- plot_lmer_gg %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
plot_lmer_gg2 <- plot_lmer_gg2 %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_mixed_tmp <- fit_mixed_tmp %>% set_names(sort(unique(NODDI_demo_scale$atlas)))
fit_effects <- names(fit_mixed_tmp) %>% map(~ emmeans::emmeans(fit_mixed_tmp[[.x]],
c("stage","ISOVF","rTMS"), infer = c(T, T)) %>%
emmeans::eff_size(sigma = sigma(fit_mixed_tmp[[.x]]),
edf = df.residual(fit_mixed_tmp[[.x]]))) %>%
set_names(names(fit_mixed_tmp))
fit_effects$DLPFC2rvmPFC contrast effect.size SE df
0 0.282652806732832 sham - 2 0.282652806732832 sham 0.1846 0.315 50.5
0 0.282652806732832 sham - 0 0.282652806732832 active -0.0818 0.595 50.5
0 0.282652806732832 sham - 2 0.282652806732832 active 1.2163 0.612 50.5
2 0.282652806732832 sham - 0 0.282652806732832 active -0.2664 0.604 51.0
2 0.282652806732832 sham - 2 0.282652806732832 active 1.0317 0.619 54.8
0 0.282652806732832 active - 2 0.282652806732832 active 1.2981 0.312 51.0
lower.CL upper.CL
-0.4483 0.817
-1.2767 1.113
-0.0125 2.445
-1.4796 0.947
-0.2081 2.271
0.6724 1.924
sigma used for effect sizes: 8.837
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 fit_effects$vmPFC2DLPFC contrast effect.size SE df
0 0.13889758195332 sham - 2 0.13889758195332 sham 0.179 0.315 50.7
0 0.13889758195332 sham - 0 0.13889758195332 active -0.260 0.602 50.7
0 0.13889758195332 sham - 2 0.13889758195332 active 1.104 0.617 50.7
2 0.13889758195332 sham - 0 0.13889758195332 active -0.438 0.610 50.8
2 0.13889758195332 sham - 2 0.13889758195332 active 0.925 0.623 54.4
0 0.13889758195332 active - 2 0.13889758195332 active 1.364 0.314 50.8
lower.CL upper.CL
-0.454 0.811
-1.468 0.949
-0.135 2.343
-1.664 0.787
-0.323 2.174
0.733 1.994
sigma used for effect sizes: 8.556
Degrees-of-freedom method: inherited from kenward-roger when re-gridding
Confidence level used: 0.95 Running all analysis and plot them
library(tidyverse)
library(ggpubr)
library(gt)
library(hrbrthemes)
library(viridis)
library(pivottabler)
#library(normwhn.test)
#library(ggiraph)
#library(ggiraphExtra)
#library(moonBook)
library(lme4)
library(car)
library(effects)
library(interactions)
#library(sjPlot)
library(lmerTest)
library(cowplot)
Fig1A <-ggdraw() + draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/E_realnorm_avg.png")
Fig1B <-ggdraw() + draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/E_realnorm_std.png")
Fig1 <- ggarrange(Fig1A,ggplot() + theme_void(),Fig1B,ncol = 3,
labels = c("A","","B"), widths = c(1,0.072,1))
ggsave(Fig1,filename = "~/Documents/Psilantro/NODDI_tms/masks/images/Figure1.png",
dpi = 300, height = 4, width = 8.5, bg = "white")
my_pal <- rcartocolor::carto_pal(n = 3, name = "Bold")
my_pal2 <- rcartocolor::carto_pal(n = 3, name = "Bold")[c(2,1,3)]
# lmm ----------------------------------------------------------
source("~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ICVF_final_Aim1B.R")
mod_fig_lm <- function(plote){
plote +
theme_bw() +
theme(text = element_text(size = 20),
legend.position="none",plot.title = element_blank(),
axis.title.x = element_blank(),axis.text.x = element_blank())+
scale_color_manual(values = my_pal2) +
scale_fill_manual(values = my_pal2)
}
plots <- list()
plots$DLPFC2rvmPFC <- mod_fig_lm(plot_lmer_probeICV$DLPFC2rvmPFC$interactplot)
plots$vmPFC2DLPFC <- mod_fig_lm(plot_lmer_probeICV$vmPFC2DLPFC$interactplot)
source("~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_OD_final_Aim1B.R")
plotsOD <- list()
plotsOD$DLPFC2Caud <- mod_fig_lm(plot_lmer_probeOD$DLPFC2Caud$interactplot)
plotsOD$DLPFC2Thal <- mod_fig_lm(plot_lmer_probeOD$DLPFC2Caud$interactplot)
plotsOD$vmPFC2DLPFC <- mod_fig_lm(plot_lmer_probeOD$vmPFC2DLPFC$interactplot) +
theme(axis.text.x = element_text(),axis.title.x = element_text()) +
scale_x_continuous(breaks=c(0,0,0,2),labels=c(" T0","","","T1 ")) +
labs(x="Time-point")
legend <- get_legend(plot_lmer_probeOD$vmPFC2DLPFC$interactplot+
scale_color_manual(values = my_pal2) +
scale_fill_manual(values = my_pal2) +
theme(legend.position="bottom",
text = element_text(size = 25)))
#----
library(cowplot)
DLPFC2rvmPFC <- ggdraw()+draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_DLPFC2rvmPFC_ls.png")
DLPFC2Thal <- ggdraw()+draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_DLPFC2Thal_ls.png")
vmPFC2DLPFC <- ggdraw()+draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_vmPFC2DLPFC_ls.png")
DLPFC2Caud <- ggdraw()+draw_image("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_DLPFC2Caud_ls.png")
brain <- ggarrange(DLPFC2rvmPFC+draw_label("DLPFC2rvmPFC",y = 0.8),
DLPFC2Thal+draw_label("DLPFC2Thal",y = 0.8),
vmPFC2DLPFC+draw_label("vmPFC2DLPFC",y = 0.8),
DLPFC2Caud+draw_label("DLPFC2Caud",y = 0.8),
ncol = 2,nrow = 2)
cuca1 <- ggarrange(brain,plots$DLPFC2rvmPFC,
plots$vmPFC2DLPFC,ncol = 3,labels = c("A","B","C"))
cuca2 <- ggarrange(plotsOD$DLPFC2Caud,plotsOD$DLPFC2Thal,
plotsOD$vmPFC2DLPFC,
ncol = 3,common.legend = TRUE,
legend = "bottom",labels = c("D","E","F"))
cuca3 <- ggarrange(cuca1,cuca2,ncol = 1, common.legend = TRUE,
legend = "bottom")
#-----------
cocoICV_fun <- function(region,lab){
sup <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/superior/tck_",region,"_s.png"))
ls <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_",region,"_ls.png"))
ant <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/ant/tck_",region,"_a.png"))
brain <- ggarrange(sup,ant,ls,ncol = 3)
brain <- ggdraw(add_sub(brain, paste0(region, " tract")))
plot_bg <- ggarrange(brain,ggplot() + theme_void(),plots[[region]],
ncol = 3,widths = c(1.7,0.042,1.2),
font.label = list(size=19),
labels = lab,vjust = -0.1)
}
cocoOD_fun <- function(region,lab){
sup <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/superior/tck_",region,"_s.png"))
ls <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/ls/tck_",region,"_ls.png"))
ant <- ggdraw()+draw_image(paste0("~/Documents/Psilantro/NODDI_tms/masks/images/ant/tck_",region,"_a.png"))
brain <- ggarrange(sup,ant,ls,ncol = 3)
brain <- ggdraw(add_sub(brain, paste0(region, " tract")))
plot_bg <- ggarrange(brain,ggplot() + theme_void(),plotsOD[[region]],
ncol = 3,widths = c(1.7,0.042,1.2),
font.label = list(size=19),
labels = lab,vjust = -0.1)
}
DLPFC2rvmPFC_ICV <- cocoICV_fun("DLPFC2rvmPFC",c("A1)","","A2)"))
vmPFC2DLPFC_ICV <- cocoICV_fun("vmPFC2DLPFC",c("B1)","","B2)"))
DLPFC2Caud_OD <- cocoOD_fun("DLPFC2Caud",c("C1)","","C2)"))
DLPFC2Thal_OD <- cocoOD_fun("DLPFC2Thal",c("D1)","","D2)"))
vmPFC2DLPFC_OD <- cocoOD_fun("vmPFC2DLPFC",c("E1)","","E2)"))
cuca <- ggarrange(ggplot() + theme_void(),ggplot() + theme_void(),legend,ncol = 3,widths = c(1.7,0.042,1.25))
cucamouse <- ggarrange(ggplot() + theme_void(),
DLPFC2rvmPFC_ICV,ggplot() + theme_void(),
vmPFC2DLPFC_ICV,ggplot() + theme_void(),
DLPFC2Caud_OD,ggplot() + theme_void(),
DLPFC2Thal_OD,ggplot() + theme_void(),
vmPFC2DLPFC_OD,cuca,
ncol = 1,heights = c(0.12,
1.4,0.12,1.4,0.12,
1.4,0.12,1.4,0.12,
1.45,0.3))
cucamouse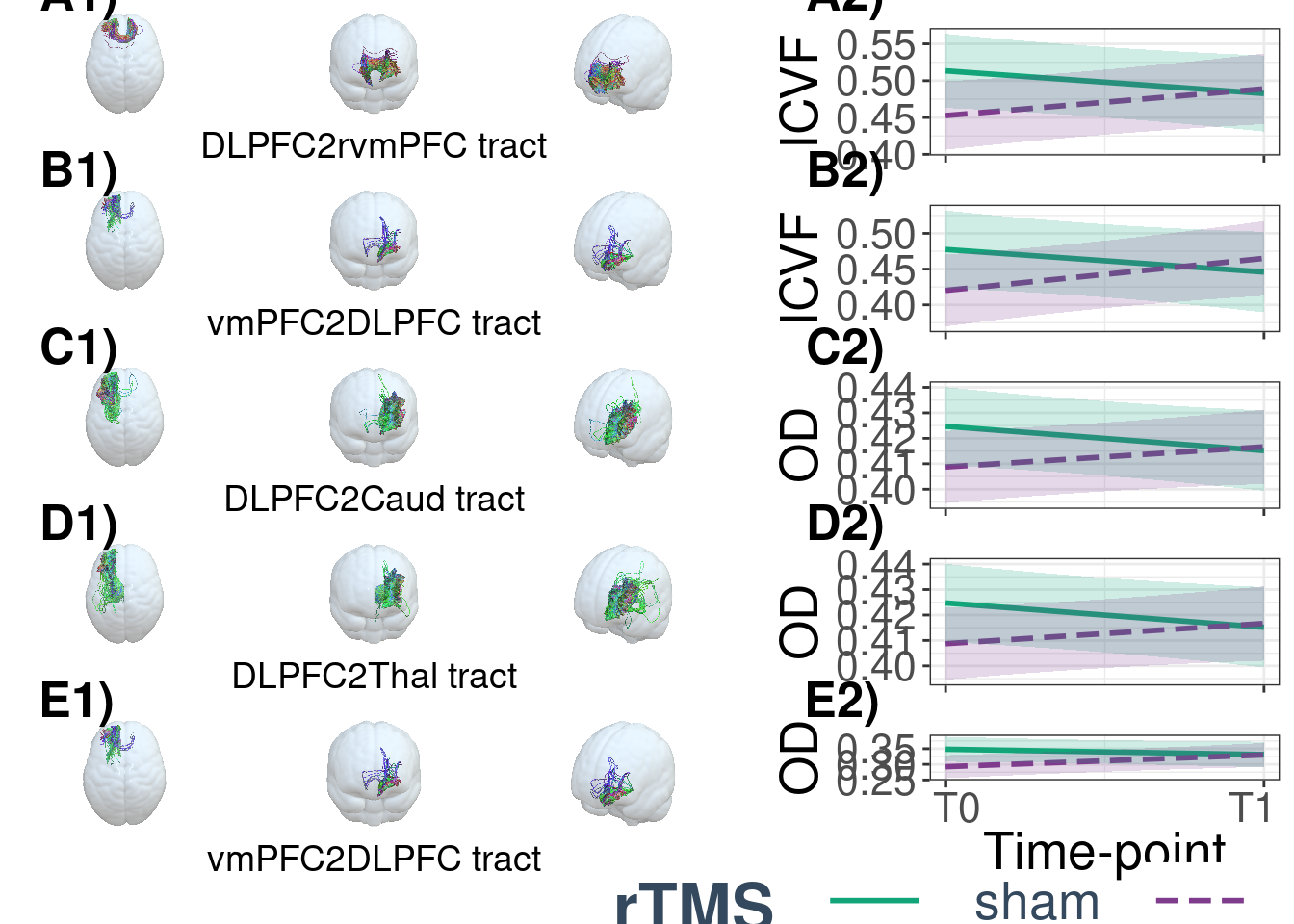
library(svglite)
svglite(filename = "~/Documents/Psilantro/NODDI_tms/masks/images/Figure2.svg",
height = 18,width = 14.5,bg = "white")
cucamouse
dev.off()png
2 ggsave(filename = "~/Documents/Psilantro/NODDI_tms/masks/images/Figure2.png",
cucamouse,dpi = 300,height = 18,width = 14.5,bg = "white",scale = 0.842)
# lmm predict ---------------------------------------------------
options(digits = 1)
source("~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ICVF_final.R")
mod_fig <- function(plot){
plot+theme(text = element_text(size = 24),
axis.text.x = element_blank(),
axis.title.y = element_text(size=18),
axis.title.x = element_text(color="white",size = 3),
plot.title = element_text(size = 18,color = "white"))+
scale_color_manual(values = my_pal) +
scale_fill_manual(values = my_pal) +
scale_x_continuous(breaks=c(0,0,0,2),labels=c(" T0","","","T1 "))+
ggtitle('VAS score predicted by ICVF in Caud2Medulla')
}
plot_lmer_gg$Caud2Medulla <- mod_fig(plot_lmer_gg$Caud2Medulla) + facet_wrap(~ICVF,nrow = 1,ncol = 5)
plot_lmer_gg$Caud2Palli <- mod_fig(plot_lmer_gg$Caud2Palli) + facet_wrap(~ICVF,nrow = 1,ncol = 5)
plot_lmer_gg$Thal2Medulla <- mod_fig(plot_lmer_gg$Thal2Medulla) + facet_wrap(~ICVF,nrow = 1,ncol = 5)
plot_lmer_gg$Thal2Palli <- plot_lmer_gg$Thal2Palli+theme(text = element_text(size = 24),
axis.title.y = element_text(size=18),
plot.title = element_text(size = 18,color = "white"))+
labs(x="Time-point")+
scale_color_manual(values = my_pal) +
scale_fill_manual(values = my_pal)+facet_wrap(~ICVF,nrow = 1,ncol = 5)+
scale_x_continuous(breaks=c(0,0,0,2),labels=c(" T0","","","T1 "))+
ggtitle('VAS score predicted by ICVF in Thal2Palli')
cucamouse_pred <- ggarrange(plot_lmer_gg$Caud2Medulla,
plot_lmer_gg$Caud2Palli,
plot_lmer_gg$Thal2Medulla,
plot_lmer_gg$Thal2Palli,
ncol = 1,nrow = 4,common.legend = T,legend = "bottom",
hjust = 0,font.label = list(size=18,face='bold'),
labels = c("A) VAS score predicted by ICVF in Caud2Medulla",
"B) VAS score predicted by ICVF in Caud2Palli",
"C) VAS score predicted by ICVF in Thal2Medulla",
"D) VAS score predicted by ICVF in Thal2Palli"))
ggsave(filename = "~/Documents/Psilantro/NODDI_tms/masks/images/Figure3-2.png",
cucamouse_pred,dpi = 300,height = 16,width = 14,bg = "white")
cucamouse_pred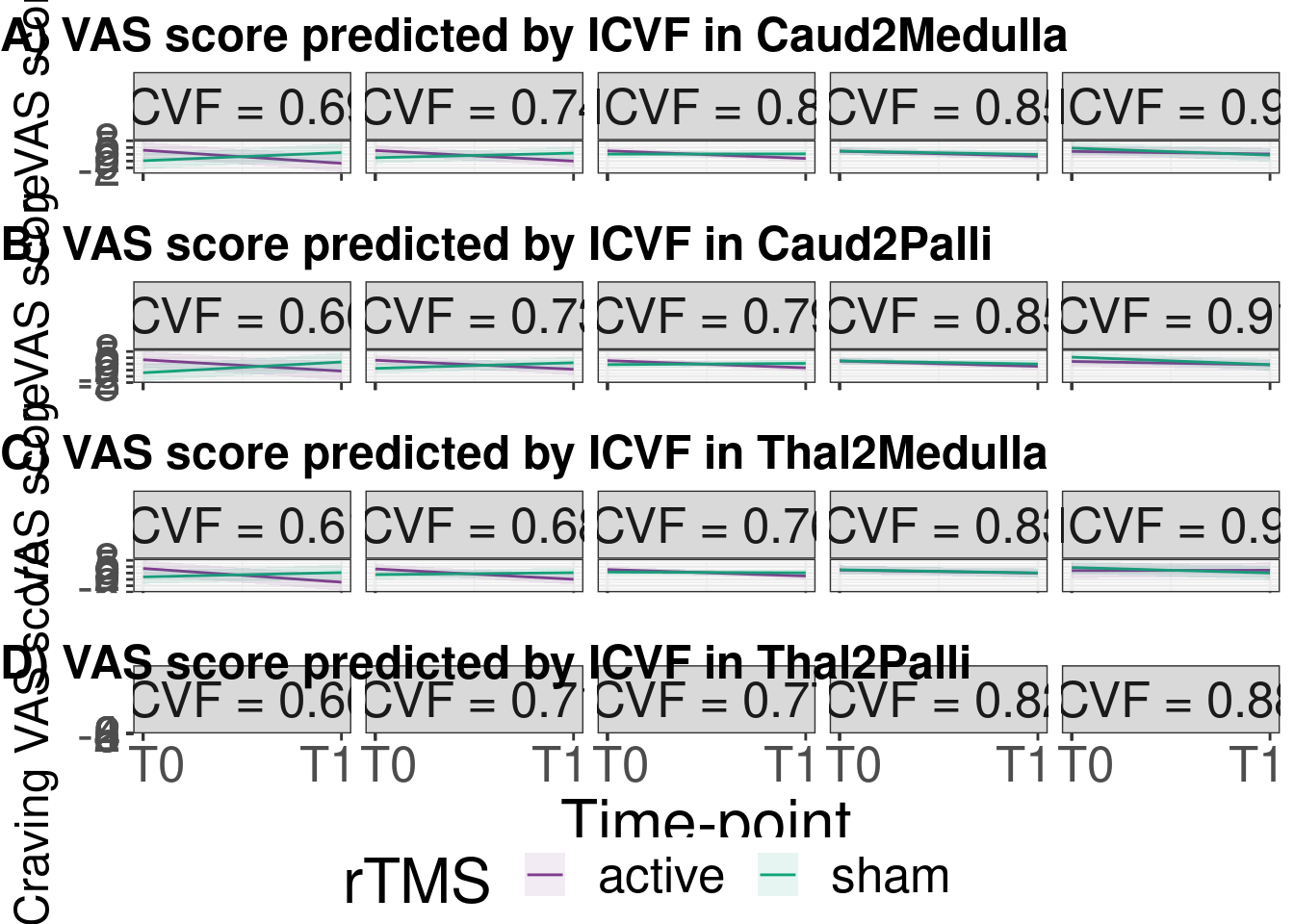
#------
source("~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_ISOVF_final.R")
plot_lmer_ggISO <- plot_lmer_gg
source("~/Documents/Psilantro/NODDI_tms/NODDI-changes-by_rTMS-in_CUD/code/analysis_OD_final.R")
plot_lmer_ggOD <- plot_lmer_gg
plot_lmer_ggISO$DLPFC2rvmPFC <- mod_fig(plot_lmer_ggISO$DLPFC2rvmPFC) + facet_wrap(~ISOVF,nrow = 1,ncol = 5)
plot_lmer_ggISO$vmPFC2DLPFC <- mod_fig(plot_lmer_ggISO$vmPFC2DLPFC) + facet_wrap(~ISOVF,nrow = 1,ncol = 5)
plot_lmer_ggOD$DLPFC2rvmPFC <- mod_fig(plot_lmer_ggOD$DLPFC2rvmPFC) + facet_wrap(~OD,nrow = 1,ncol = 5) +
theme(axis.text.x = element_text())
cucamouse_pred2 <- ggarrange(plot_lmer_ggISO$DLPFC2rvmPFC,
plot_lmer_ggISO$vmPFC2DLPFC,
plot_lmer_ggOD$DLPFC2rvmPFC,
ncol = 1,nrow = 3,common.legend = T,legend = "bottom",
hjust = 0,font.label = list(size=18,face='bold'),
labels = c("A) BIS-11 score predicted by ISOVF in DLPFC2rvmPFC",
"B) BIS-11 score predicted by ISOVF in vmPFC2DLPFC",
"C) BIS-11 score predicted by OD in DLPFC2rvmPFC"))
ggsave(filename = "~/Documents/Psilantro/NODDI_tms/masks/images/Figure4-2.png",
cucamouse_pred2,dpi = 300,height = 12,width = 14,bg = "white")
cucamouse_pred2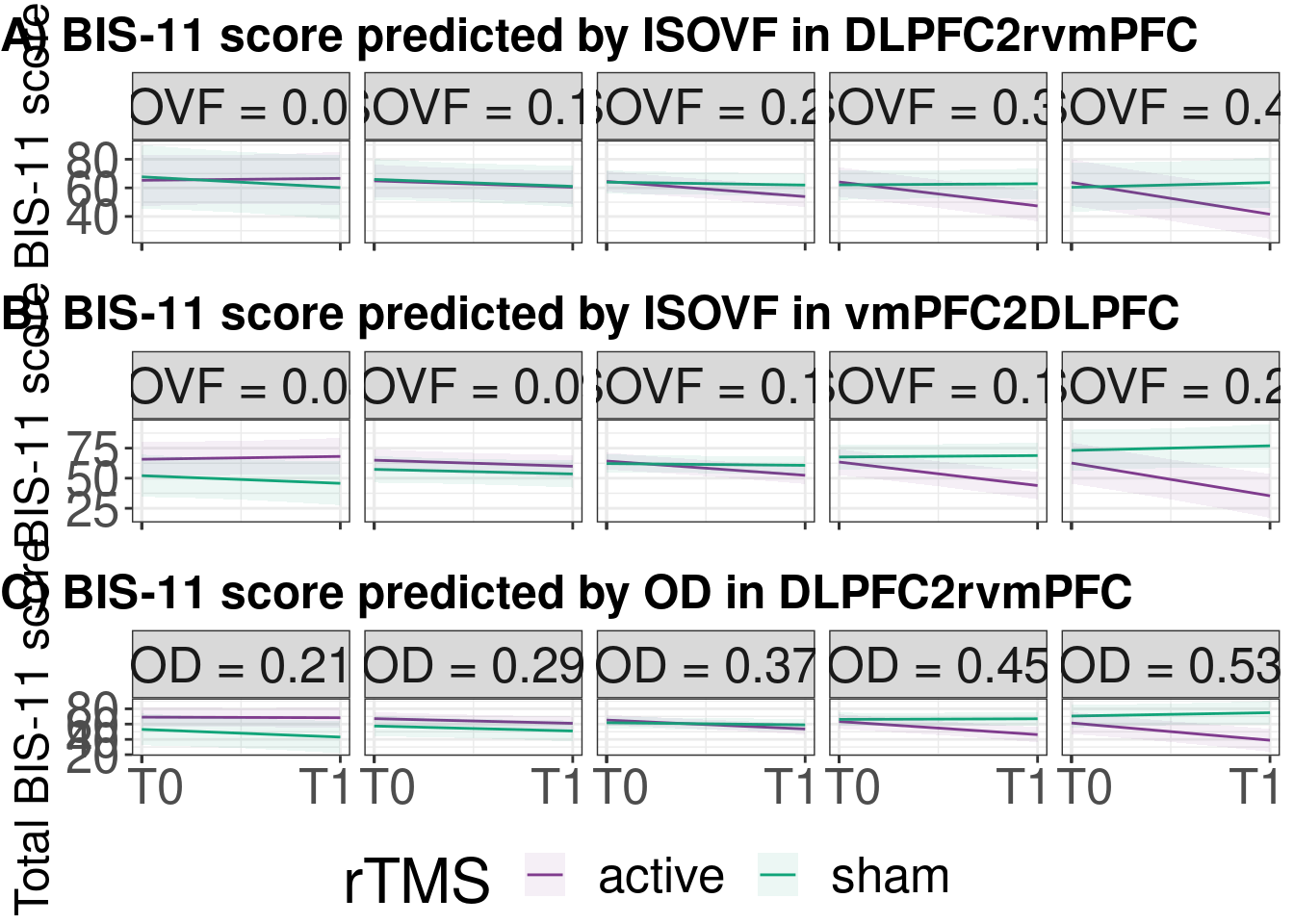
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.5 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=es_MX.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=es_MX.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=es_MX.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=es_MX.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] svglite_2.1.0 cowplot_1.1.1 lmerTest_3.1-3 interactions_1.1.5
[5] effects_4.2-1 car_3.1-0 carData_3.0-5 lme4_1.1-30
[9] Matrix_1.5-1 pivottabler_1.5.3 viridis_0.6.2 viridisLite_0.4.1
[13] hrbrthemes_0.8.0 gt_0.7.0 ggpubr_0.4.0 forcats_0.5.2
[17] stringr_1.4.1 dplyr_1.0.9 purrr_0.3.4 readr_2.1.2
[21] tidyr_1.2.0 tibble_3.1.8 ggplot2_3.3.6 tidyverse_1.3.1
[25] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] readxl_1.4.1 backports_1.4.1 jtools_2.1.4
[4] systemfonts_1.0.4 splines_4.1.2 TH.data_1.1-0
[7] digest_0.6.29 htmltools_0.5.3 magick_2.7.3
[10] fansi_1.0.3 magrittr_2.0.3 tzdb_0.3.0
[13] modelr_0.1.8 extrafont_0.18 vroom_1.5.7
[16] sandwich_3.0-1 extrafontdb_1.0 colorspace_2.0-3
[19] rvest_1.0.2 mitools_2.4 textshaping_0.3.6
[22] haven_2.5.1 xfun_0.32 callr_3.7.2
[25] crayon_1.5.1 jsonlite_1.8.0 survival_3.3-1
[28] zoo_1.8-10 glue_1.6.2 gtable_0.3.0
[31] emmeans_1.7.3 Rttf2pt1_1.3.10 abind_1.4-5
[34] scales_1.2.1 mvtnorm_1.1-3 DBI_1.1.2
[37] rstatix_0.7.0 ggthemes_4.2.4 Rcpp_1.0.9
[40] xtable_1.8-4 bit_4.0.4 survey_4.1-1
[43] htmlwidgets_1.5.4 httr_1.4.2 ellipsis_0.3.2
[46] farver_2.1.1 pkgconfig_2.0.3 nnet_7.3-17
[49] sass_0.4.2 dbplyr_2.1.1 utf8_1.2.2
[52] labeling_0.4.2 tidyselect_1.1.2 rlang_1.0.4
[55] later_1.3.0 munsell_0.5.0 cellranger_1.1.0
[58] tools_4.1.2 cachem_1.0.6 cli_3.3.0
[61] generics_0.1.3 broom_1.0.0 evaluate_0.16
[64] fastmap_1.1.0 ragg_1.2.2 yaml_2.3.5
[67] processx_3.7.0 knitr_1.40 bit64_4.0.5
[70] fs_1.5.2 pander_0.6.5 nlme_3.1-155
[73] whisker_0.4 xml2_1.3.3 compiler_4.1.2
[76] pbkrtest_0.5.1 rstudioapi_0.14 ggsignif_0.6.3
[79] reprex_2.0.1 bslib_0.4.0 stringi_1.7.8
[82] highr_0.9 ps_1.7.1 gdtools_0.2.4
[85] lattice_0.20-45 nloptr_1.2.2.3 vctrs_0.4.1
[88] pillar_1.8.1 lifecycle_1.0.1 jquerylib_0.1.4
[91] estimability_1.3 data.table_1.14.2 insight_0.18.2
[94] rcartocolor_2.0.0 httpuv_1.6.5 R6_2.5.1
[97] promises_1.2.0.1 gridExtra_2.3 codetools_0.2-18
[100] boot_1.3-28 MASS_7.3-58.1 assertthat_0.2.1
[103] rprojroot_2.0.3 withr_2.5.0 multcomp_1.4-18
[106] parallel_4.1.2 hms_1.1.2 grid_4.1.2
[109] coda_0.19-4 minqa_1.2.4 rmarkdown_2.16
[112] git2r_0.30.1 getPass_0.2-2 numDeriv_2016.8-1.1
[115] lubridate_1.8.0